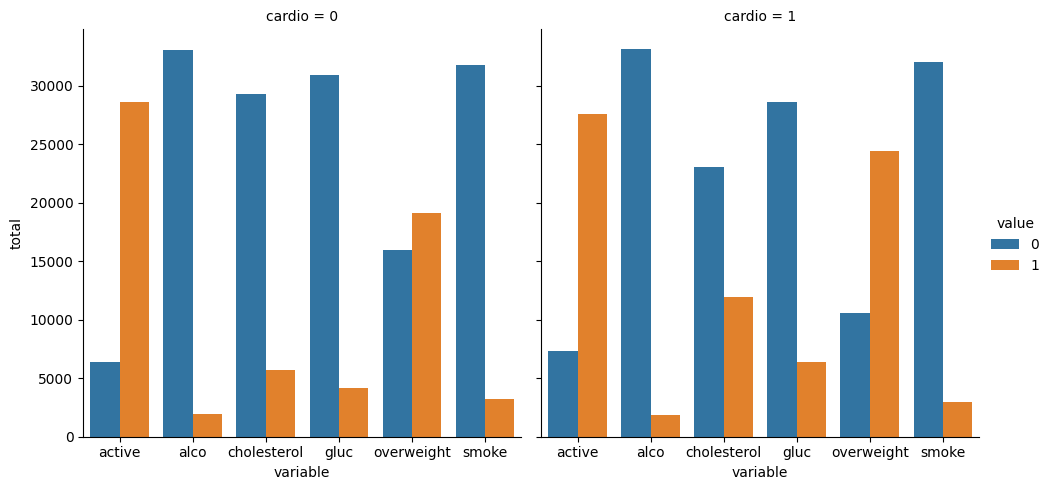
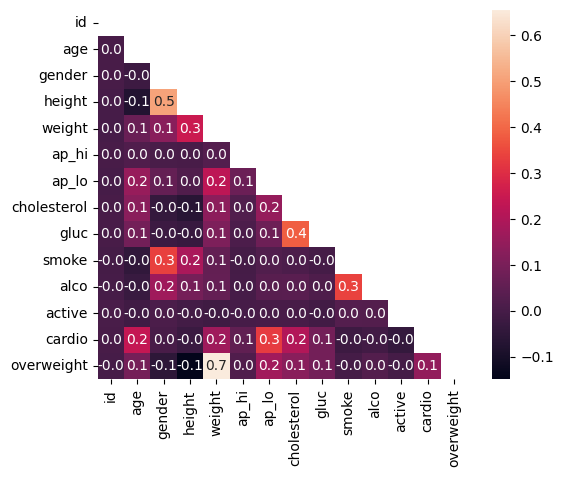
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
# Import data
df = pd.read_csv("data/medical_examination.csv")
df| id | age | gender | height | weight | ap_hi | ap_lo | cholesterol | gluc | smoke | alco | active | cardio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 18393 | 2 | 168 | 62.0 | 110 | 80 | 1 | 1 | 0 | 0 | 1 | 0 |
| 1 | 1 | 20228 | 1 | 156 | 85.0 | 140 | 90 | 3 | 1 | 0 | 0 | 1 | 1 |
| 2 | 2 | 18857 | 1 | 165 | 64.0 | 130 | 70 | 3 | 1 | 0 | 0 | 0 | 1 |
| 3 | 3 | 17623 | 2 | 169 | 82.0 | 150 | 100 | 1 | 1 | 0 | 0 | 1 | 1 |
| 4 | 4 | 17474 | 1 | 156 | 56.0 | 100 | 60 | 1 | 1 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 69995 | 99993 | 19240 | 2 | 168 | 76.0 | 120 | 80 | 1 | 1 | 1 | 0 | 1 | 0 |
| 69996 | 99995 | 22601 | 1 | 158 | 126.0 | 140 | 90 | 2 | 2 | 0 | 0 | 1 | 1 |
| 69997 | 99996 | 19066 | 2 | 183 | 105.0 | 180 | 90 | 3 | 1 | 0 | 1 | 0 | 1 |
| 69998 | 99998 | 22431 | 1 | 163 | 72.0 | 135 | 80 | 1 | 2 | 0 | 0 | 0 | 1 |
| 69999 | 99999 | 20540 | 1 | 170 | 72.0 | 120 | 80 | 2 | 1 | 0 | 0 | 1 | 0 |
70000 rows × 13 columns